Most basic Heatmap
How to do it: below is the most basic heatmap you can build in base R, using the heatmap() function with no parameters. Note that it takes as input a matrix. If you have a data frame, you can convert it to a matrix with as.matrix(), but you need numeric variables only.
How to read it: each column is a variable. Each observation is a row. Each square is a value, the closer to yellow the higher. You can transpose the matrix with t(data) to swap X and Y axis.
Note: as you can see this heatmap is not very insightful: all the variation is absorbed by the hp and disp variables that have very high values compared to the others. We need to normalize the data, as explained in the next section.
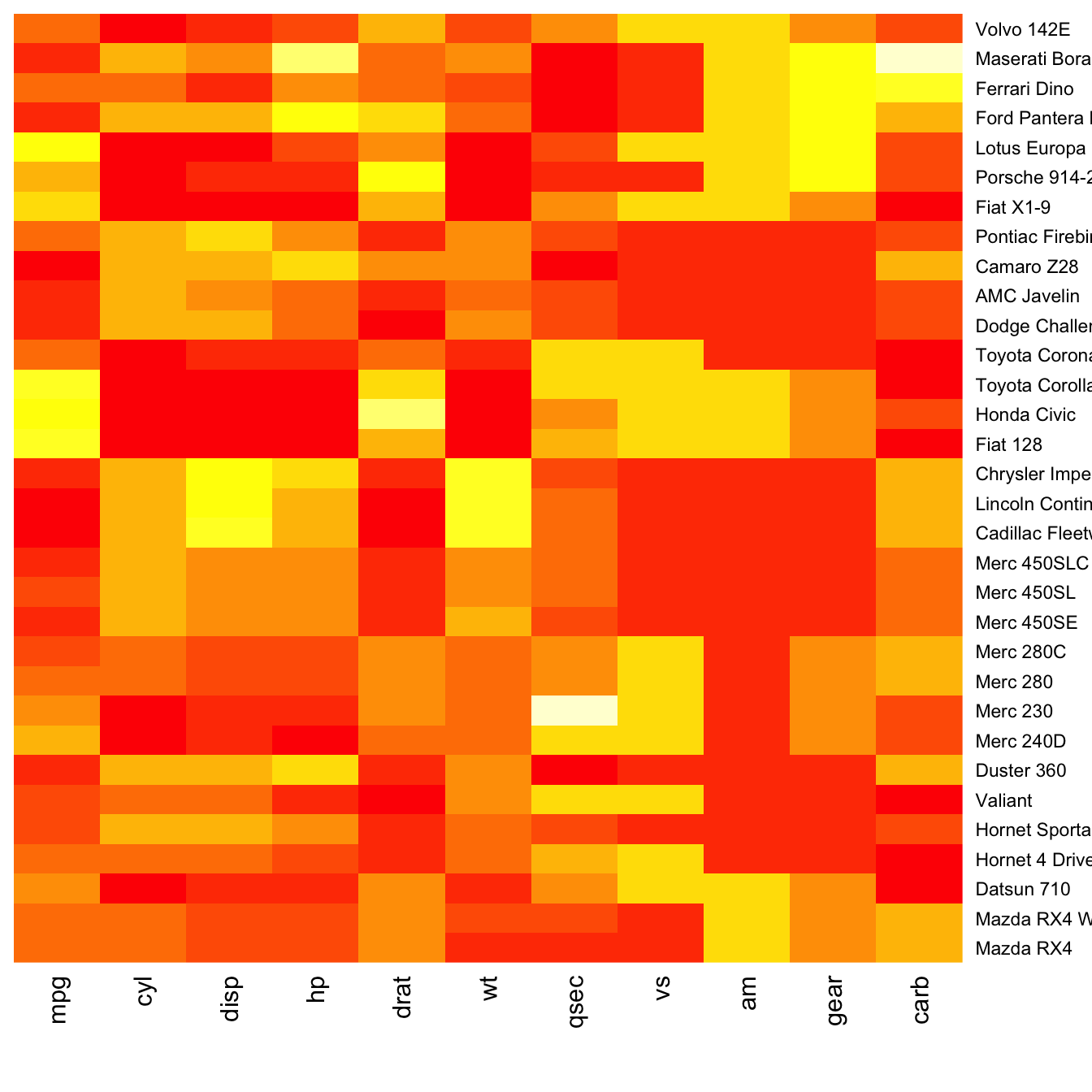
Normalization
Normalizing the matrix is done using the scale argument of the heatmap() function. It can be applied to row or to column. Here the column option is chosen, since we need to absorb the variation between column.
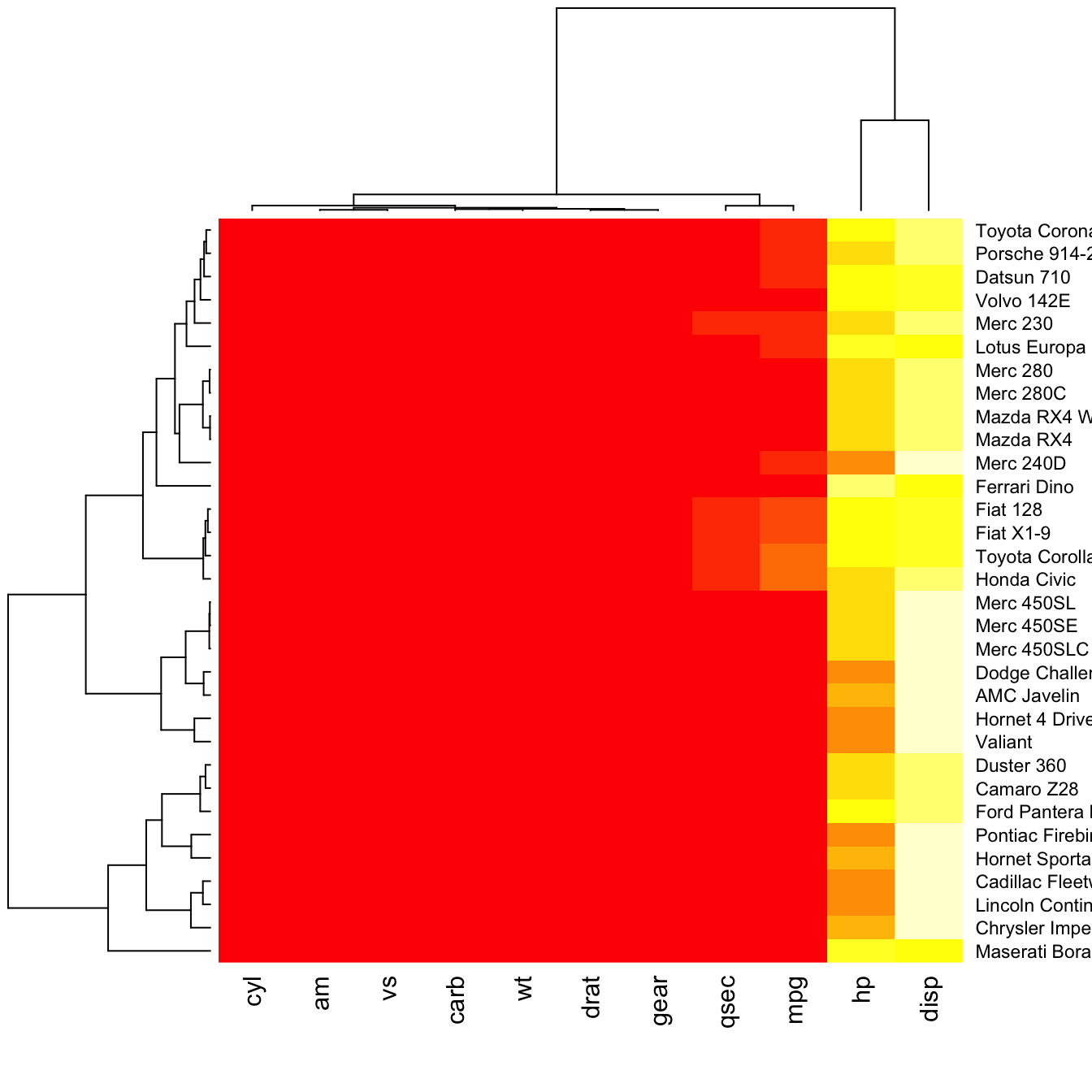
Dendrogram and Reordering
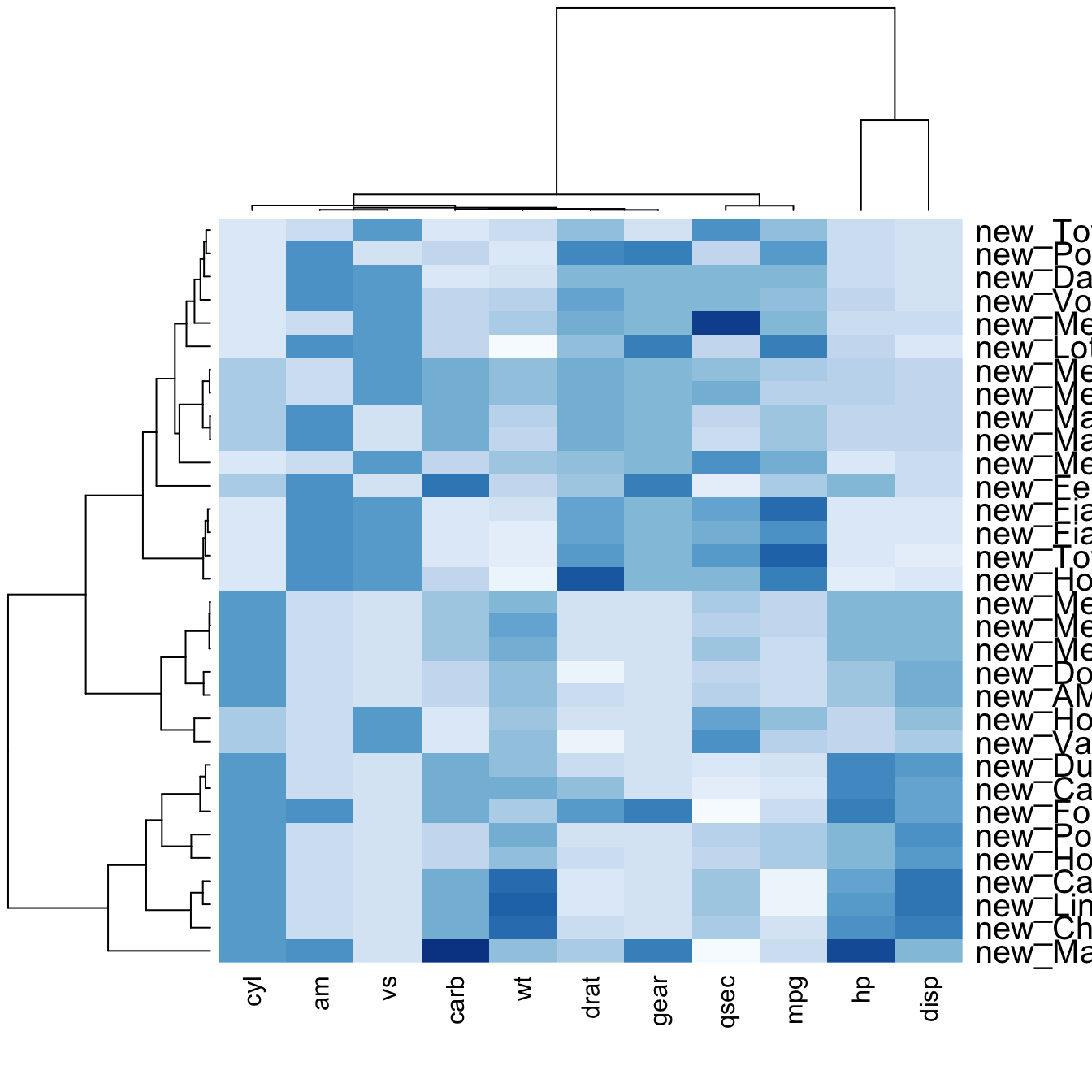
You may have noticed that order of both rows and columns is different compare to the native mtcar matrix. This is because heatmap() reorders both variables and observations using a clustering algorithm: it computes the distance between each pair of rows and columns and try to order them by similarity.
Moreover, the corresponding dendrograms are provided beside the heatmap. We can avoid it and just visualize the raw matrix: use the Rowv and Colv arguments as follow.
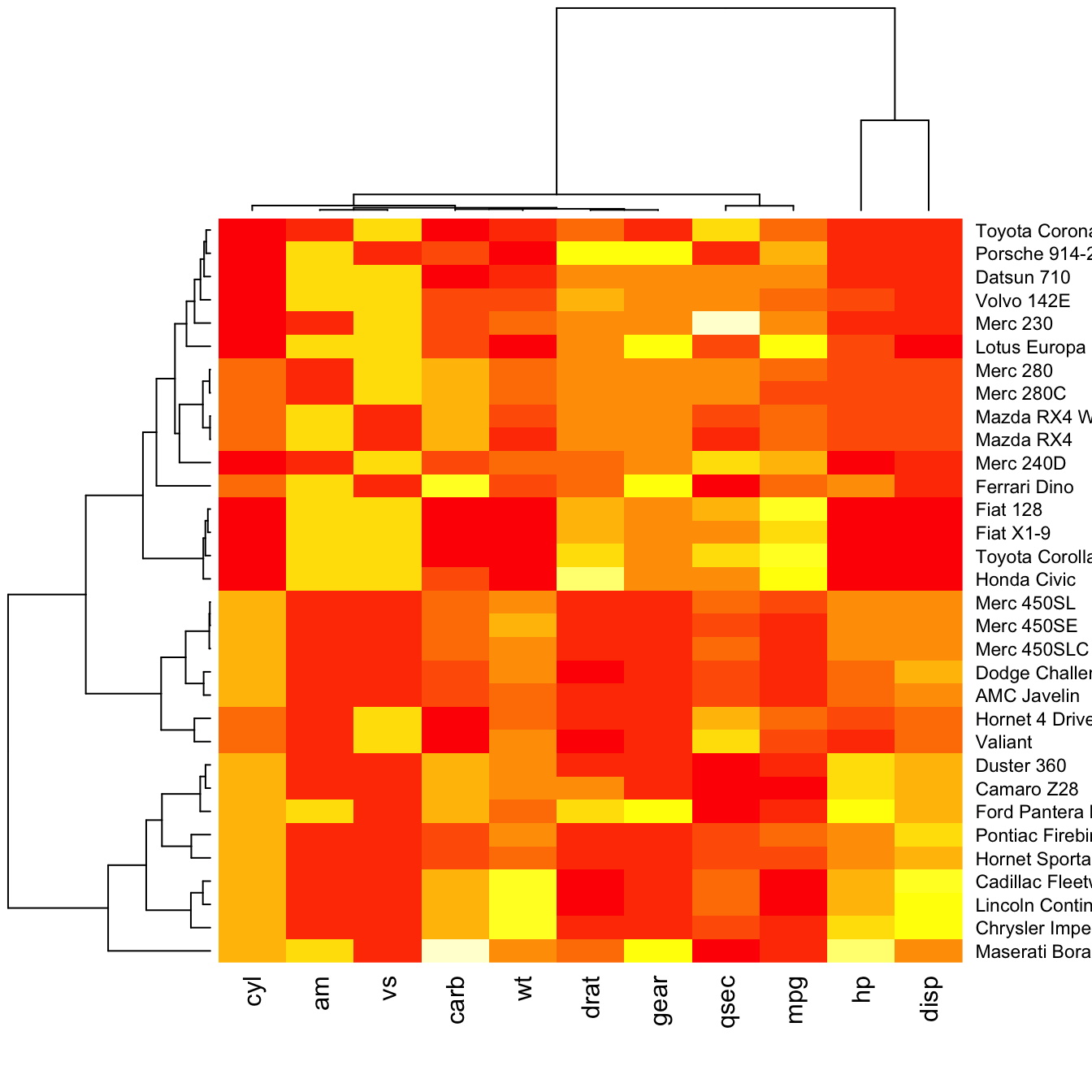
# No dendrogram nor reordering for neither column or row
heatmap(data, Colv = NA, Rowv = NA, scale="column")Color palette
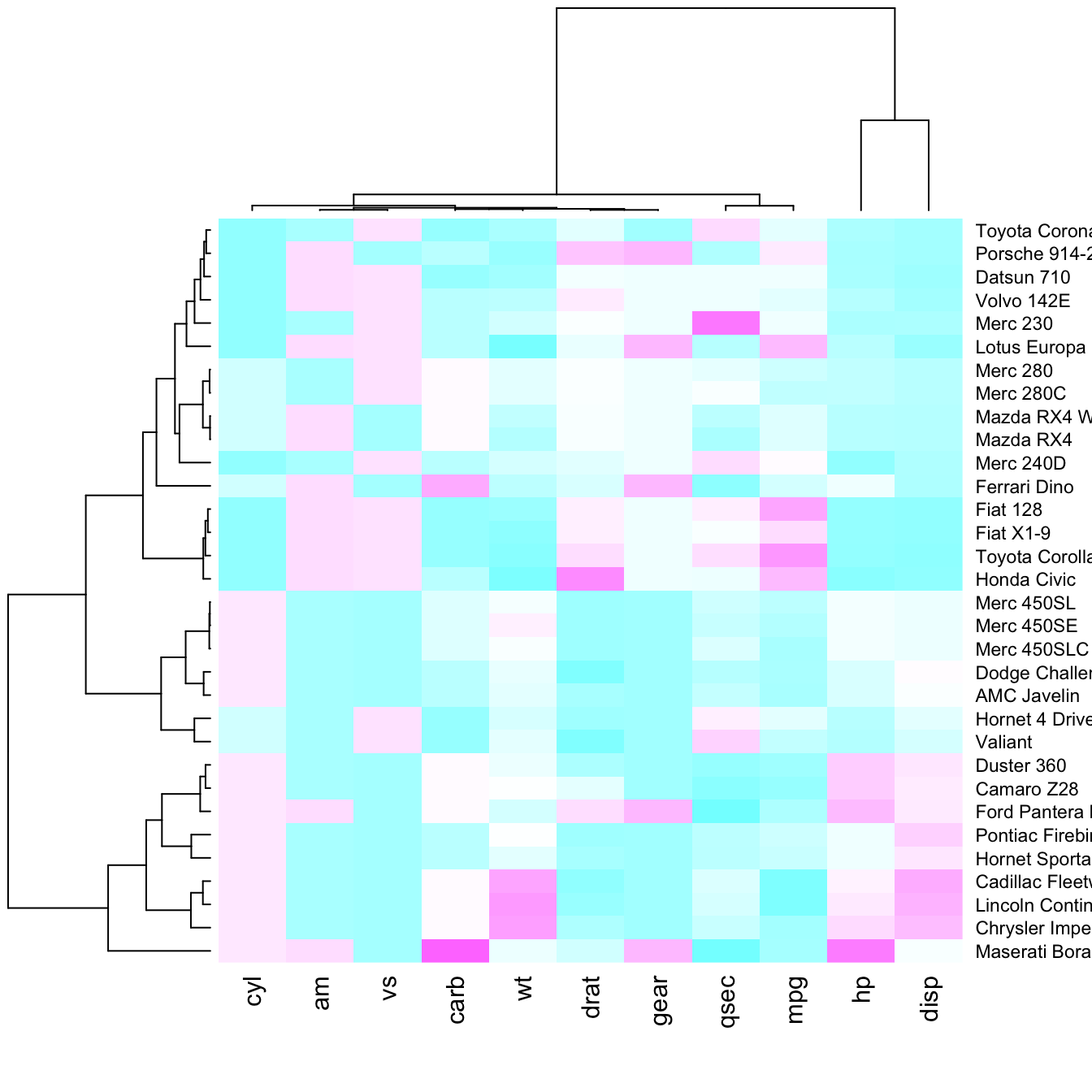
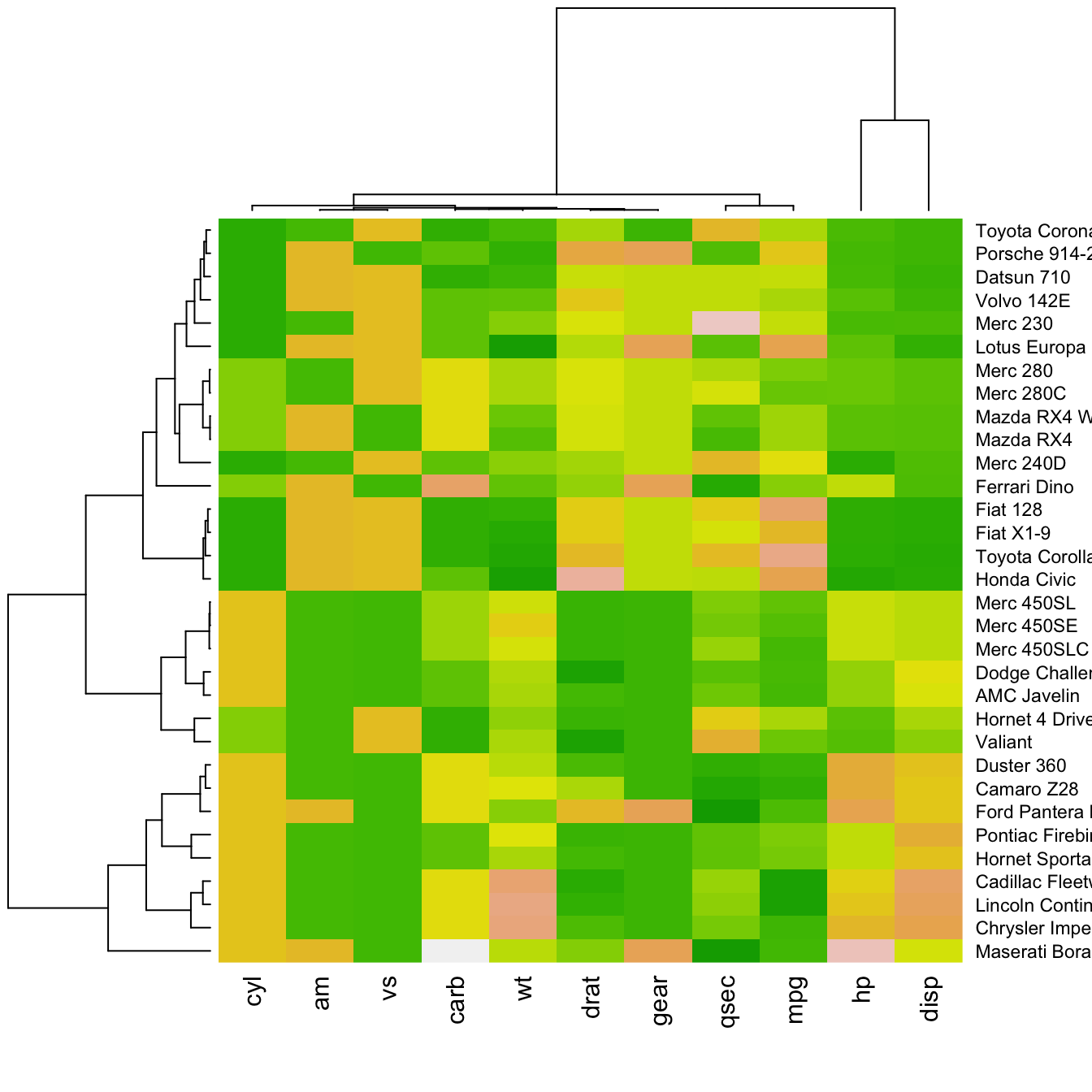
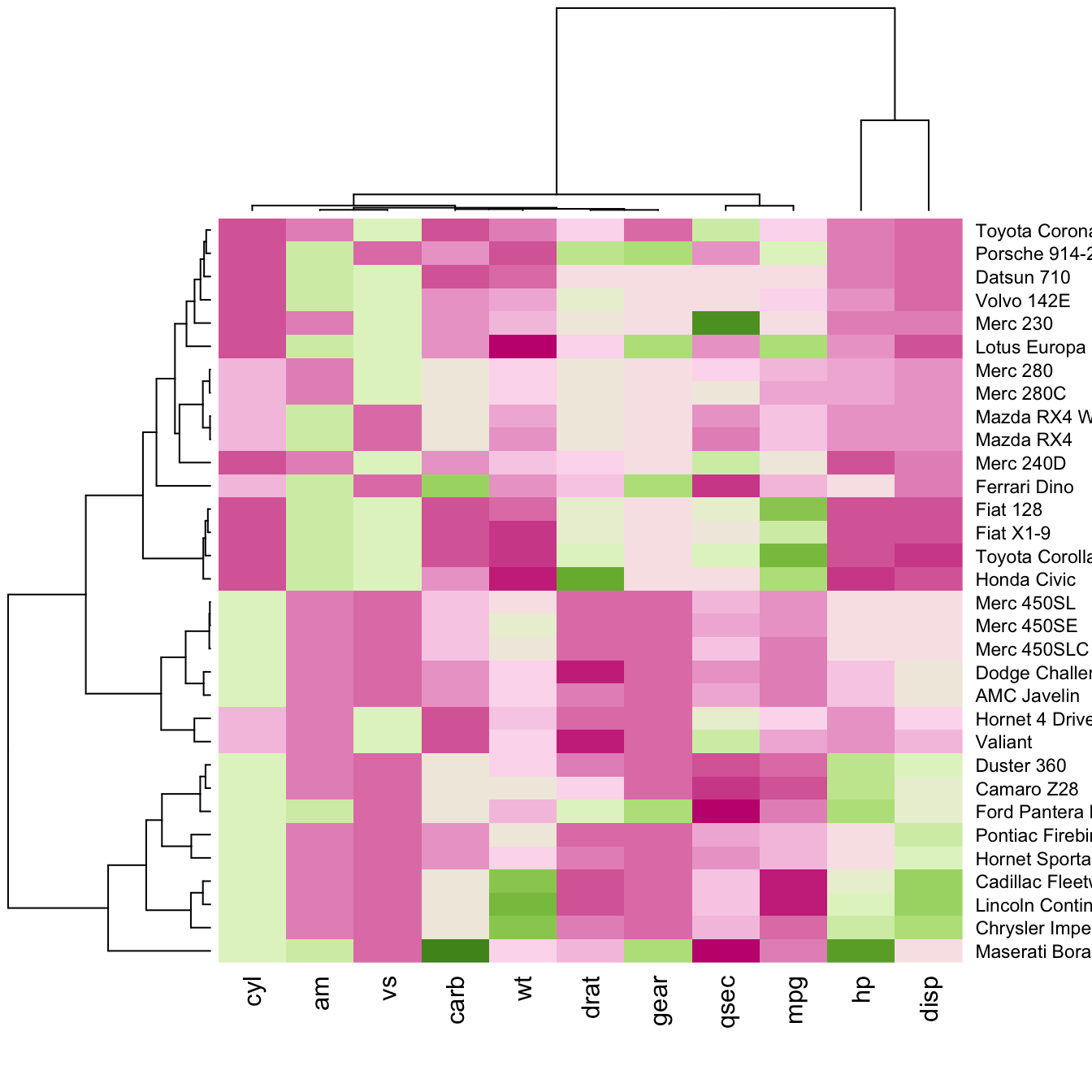
There are several ways to custom the color palette:
- use the native palettes of R:
terrain.color(),rainbow(),heat.colors(),topo.colors()orcm.colors() - use the palettes proposed by
RColorBrewer. See list of available palettes here.
Custom Layout
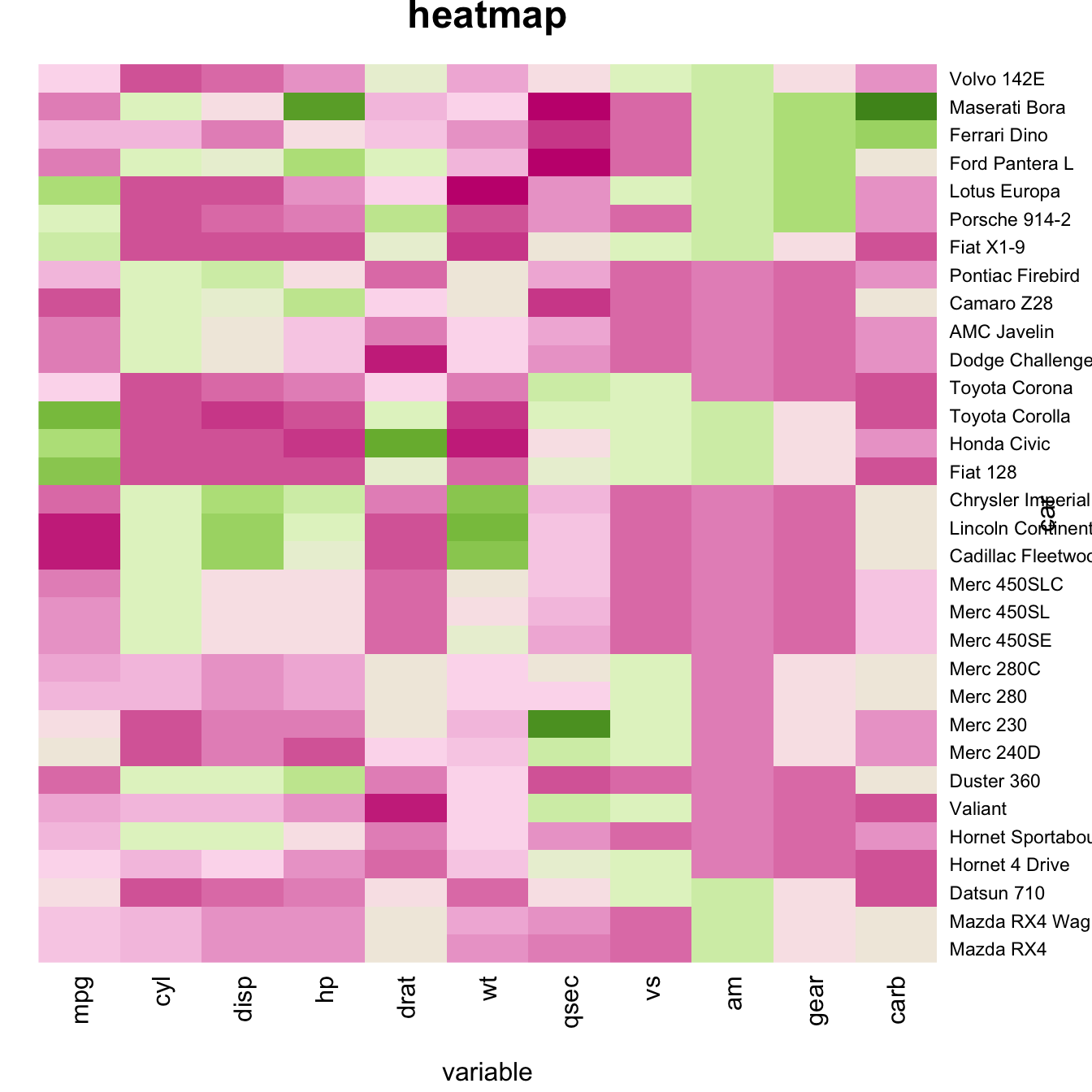
You can custom title & axis titles with the usual main and xlab/ylab arguments (left).
You can also change labels with labRow/colRow and their size with cexRow/cexCol.
# Add classic arguments like main title and axis title
heatmap(data, Colv = NA, Rowv = NA, scale="column", col = coul, xlab="variable", ylab="car", main="heatmap")
# Custom x and y labels with cexRow and labRow (col respectively)
heatmap(data, scale="column", cexRow=1.5, labRow=paste("new_", rownames(data),sep=""), col= colorRampPalette(brewer.pal(8, "Blues"))(25))Add color beside heatmap
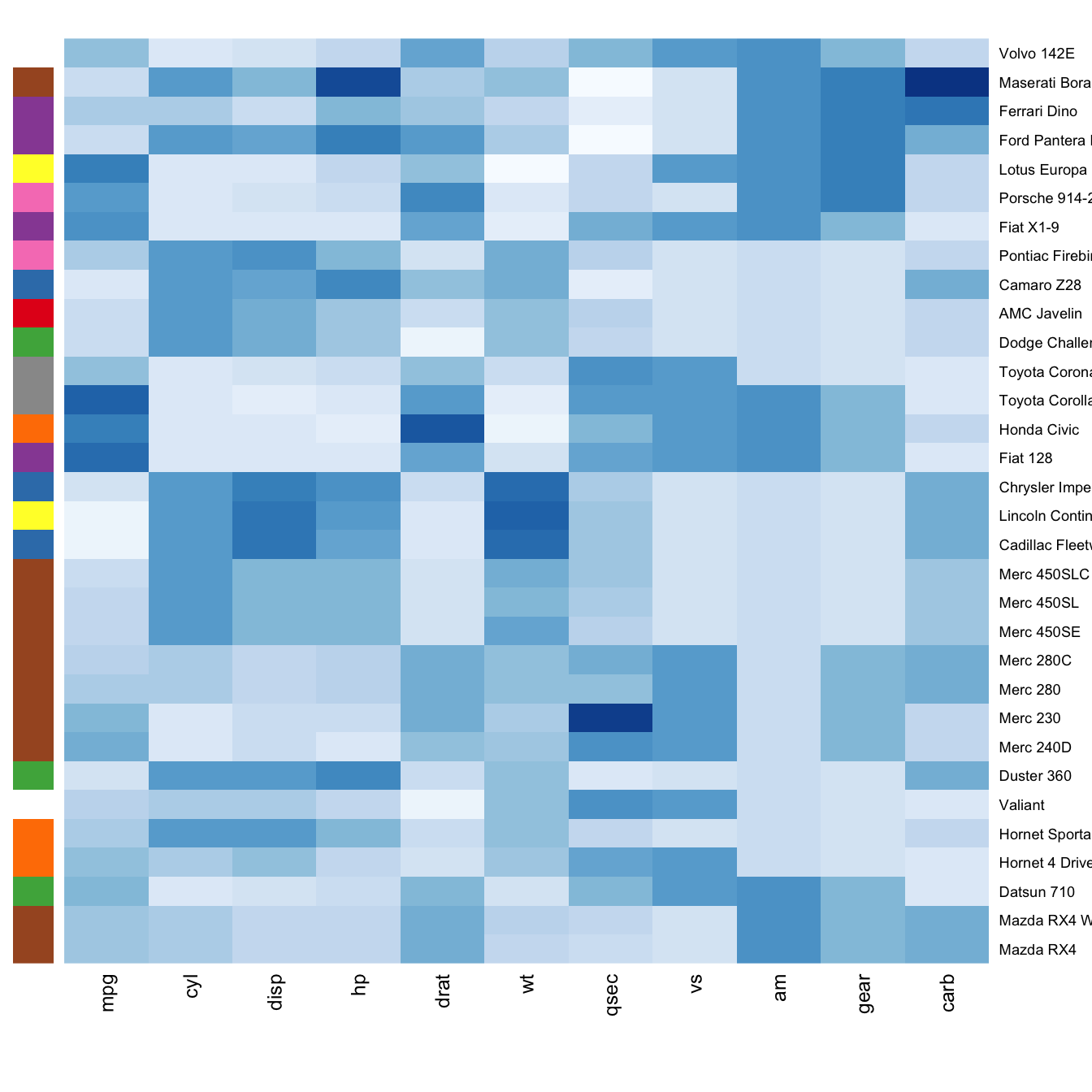
Often, heatmap intends to compare the observed structure with an expected one.
You can add a vector of color beside the heatmap to represents the expected structure using the RowSideColors argument.